Introduction
Let’s step into the world of cutting-edge technology and explore how image segmentation is revolutionizing the field of biomedicine!
Image segmentation in the biomedical field is crucial for a number of reasons. It helps to identify and separate different structures or objects within an image, which is essential for accurate diagnosis and treatment planning. With the help of image segmentation, medical practitioners can precisely isolate and analyze specific areas of interest within an image, leading to improved understanding of the underlying anatomy and disease processes. The use of deep learning algorithms in image segmentation in the biomedical field has improved the accuracy and speed of image analysis, making it a valuable tool for medical professionals.
In this article, we’ll explore the amazing nuances of one of the most popular models for deep learning-based biomedical image segmentation, UNet, and how it’s revolutionizing the medical field. So grab a coffee and let’s jump right in!
What is UNet?
UNet is a popular deep learning architecture that is widely used in image segmentation. The UNet model has been specifically designed to address the challenges of biomedical image segmentation and has achieved remarkable results in several real-life applications.
UNet was first introduced in 2015 by Olaf Ronneberger et al. in “U-Net: Convolutional Networks for Biomedical Image Segmentation.” Since it’s inception, there have been several iterations UNet++, 3D UNet, Transformer-UNet, and now UNet 3+, including plug-ins for non-machine learning experts such as this ImageJ. It’s not surprising that many improvements and adaptations have followed this popular model since it’ won the Grand Challenge for Computer-Automated Detection of Caries in Bitewing Radiography at ISBI 2015, and the Cell Tracking Challenge at ISBI 2015. For image segmentation, UNet is one of the best artificial neural network choices out there.
UNet is…
+ An end-to-end deep convolutional neural network
+ Designed for image segmentation tasks
+ Used in several real-world applications
UNet excels at…
+ Small datasets of labled biomedical data (~30)
+ Fast analysis (<1 sec.)
+ Segmentation when objects are touching
This article will provide an overview of UNet, including its architecture and its relation to deep learning.
Also Read: The role of AI in vaccine distribution
Understanding UNet
One of the problems in biomedical image segmentation is that gathering data is often expensive. X-ray images, pancreas images, 2-photon images, etc. all use quite expensive equipment and overhead, so gather the amount of data needed for most deep learning models becomes a financial challenge.In image segmentation, you take in some raw image data and try to locate by outline or highlighting an important part of the image that you’re looking for. For example, highlighting cancer in lung x-rays.
UNet solves this problem by creating an information pipeline that contracts and then expands. The image is fed into the network which then computes the segmentation map (i.e. outline or highlited region corresponding to the targets in the input).
The UNet architecture is designed to preserve the spatial information of the original image while reducing the spatial resolution of the features through average pooling and strided convolution operations. This is another useful feature of UNet since biomedical images are often very high resolution, making them less computationally expensive to analyze.
Another cool feature of UNet is that it is designed to handle differences in target boundaries and changeable targets. Which makes it a great option for biomedical images where there are many irregularities in the data.
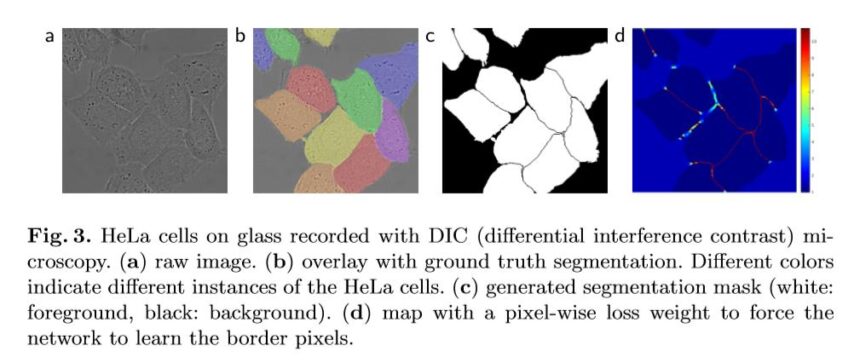
Image of various biomedical image segmentations from the original UNet paper by Ronneberger, 2015.
What is the UNet Model?
The UNet model is what we call a deep fully connected convolutional neural network. It consists of an encoder network and a decoder network. The encoder network is responsible for extracting features from the input image, while the decoder network is responsible for reconstructing the output layer segmentation map.The encoder and decoders are comprised of a series of convolutional operations with weights representing multi-channel feature maps.
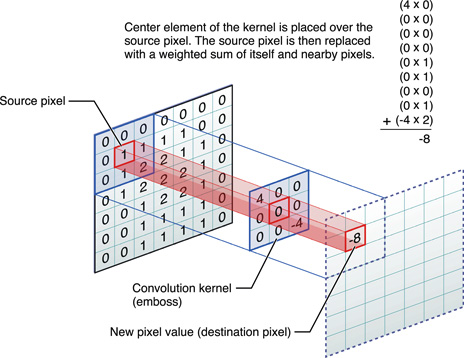
If you’re not familar with the convolution process, it involves sliding a small window, called a convolutional kernel or filter, over the image to perform a set of mathematical operations at each position. The output of the convolution operation is a transformed image that has been filtered to highlight discriminative features or patterns. In neural networks, the image pixel space is fed through convolutions and are often followed by a nonlinear activation, which is a function that introduces non-linearity, allowing the network to model complex relationships and speed up training.
UNet Architecture
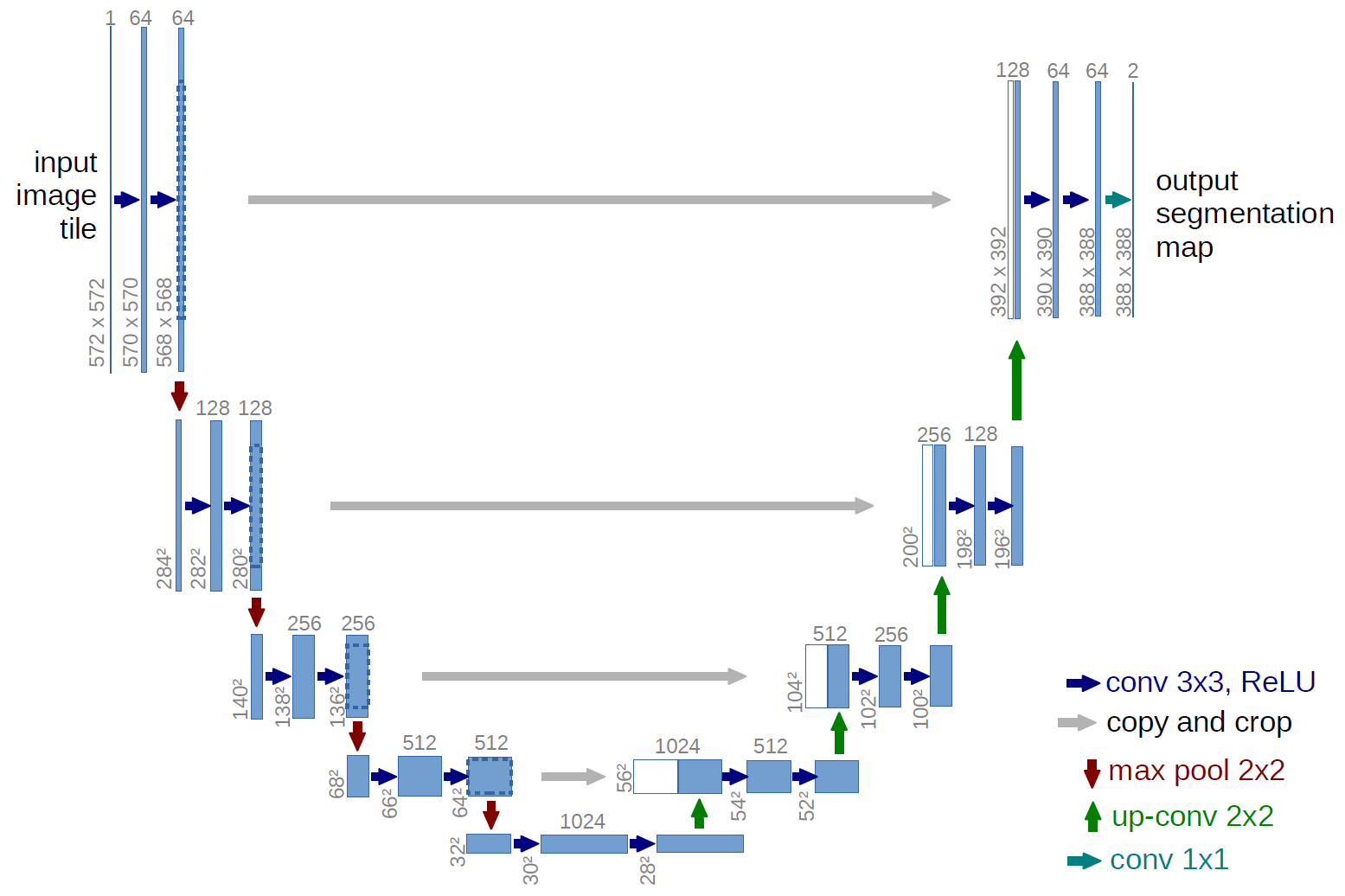
UNet’s architecture consists of a series of convolution blocks and includes multiple convolution layers, activation functions, and pooling layers. The UNet architecture also includes a depth-wise separable convolution that enables it to process images with high spatial resolution. UNet is trained using a hybrid loss function, which combines a cross-entropy loss function and a boundary-aware loss function.
Below is a diagram which shows the general components of the UNet architecture, taken from University of Freiburg, Dept. of Computer Science, by Olaf Ronneberger.
Although the architecture may look complicated, it is actually fairly simple. Below I’ll explain the encoder-decoder structure.
The Encoder
Images are reduced using smaller and smaller adjacent convolutional blocks. The input image turns into an output feature map after one convolutional block, which becomes the input feature map to the next block.
UNet’s encoder uses atrous convolutions, also known as dilated convolutions which are used to increase the resolution of feature maps, allowing for more detailed segmentation of images. This is achieved by increasing the effective field of view of the convolutional kernels, effectively increasing the coverage of the feature maps. By combining the use of pooling and atrous convolution layers, UNet can effectively balance the resolution and abstraction of the feature maps, enabling more accurate image segmentation.
The calculation of convolutions is performed as channel convolutions, or depth-wise convolutions. Each input channel is convolved with separate filters and the channel features are combined to form the output channels. This allows the network to learn complex relationships between multiple channels of information in an image, such as color and texture. The principal behind this is known as convolution for fusion where there is use of convolutional layers to combine features from multiple scales or resolutions of an image. This fusion process helps UNet capture both high-level and low-level features in an image, which is crucial for accurate image segmentation.
In order to keep lower memory requirements and faster convergance, UNet models sometimes use something called depth-separable convolution modules. In a depth-separable convolution, the spatial convolution operation and the pointwise convolution operation are separated and performed independently. This allows for a reduction in the number of convolution operator parameters required, making the model lighter and faster to train. For example, the dilated rate is a hyperparameter that determines the spacing between the values in the input that are used in the calculation of each output value. In other words, it controls the “receptive field” of the convolution operation.
After each convolution, there is an additional structure comprised of an average pooling layer is commonly used in the downsampling path (encoder) of the UNet architecture to reduce the spatial resolution of the feature maps. By taking the average of multiple neighboring pixels, the average pooling layer reduces the amount of computation needed in the subsequent layers and reduces overfitting. A spatial pyramid pooling layer is also added to capture context information at multiple scales.
Following the pooling layer, there is a residual structure designed to address the vanishing gradients problem in deep neural networks by allowing the inputs to be directly added to the outputs of each layer in the network. This helps to ensure that the network can learn to make small adjustments to the inputs rather than having to learn the entire mapping from scratch, which can be difficult for very deep networks.
The Decoder
Essentially the decoder goes through the same operrations as the encoder in reverse with a few modifications. The decoder upsamples using bilinear interpolation by estimating the values of missing pixels in an image to construct the segmentation map. This gives UNet discrimination at pixel level, increasing accuracy.
Overall
The distribution of pixel values in an image can affect the performance of a UNet model in biomedical image segmentation. A well-designed UNet should be able to handle different types of images with varying distributions of pixel values, such as images with Gaussian, Poisson, or exponential distributions.
How does it relate to Deep Learning?
UNet is itself a deep learning model. The UNet model is an example of how deep learning can be used for image segmentation tasks. For Ronneberger’s original UNet paper being cited over 56,000 times, it should come as no surprise that several models have adopted a UNet architecture as the backbone network.
One interesting approach is that of Pix2Pix, a generative adversarial network (GAN). GANs generate data to some target distribution using a generator and an adversarial model. The generator in Pix2Pix is built on top of the UNet architecture since the segmentation map is adept at fluctuating morphological features in the image.
UNet architecture is often used as the base network in several transformer models (a popular natural language processing frameworks… similar to the backbone of ChatGPT) for image segmentation tasks in the biomedical field. Some examples of such transformer models include:
- TransUNet – This is a variant of UNet that uses a transformer network in place of the usual convolutional layers in the UNet architecture.
- Attention-UNet – This is another variant that uses attention mechanisms in addition to the UNet architecture to better capture long-range dependencies in the image.
- TernausNetV2 – This is a yet another variant of the UNet architecture for satellite image detection that uses skip connections to extract targets on an instance level.
As listed above, there are several iterations of the base UNet architecture. Common improvements on UNet include:
- 3-D Fully Convolutional Networks
- dual-channel neural network
- gray level co-occurrence matrix preprocessing
Applications of UNet in real life
As you might have guessed by now, UNet has been applied to several real-life applications.
Below is a list of just a few applications.

Biomedical Image Segmentation Datsets and Tasks:

Other Computer Vision Tasks:
- Enhance sharpness of image edges in 2D images
- Remote sensing images (satellite images)
- Data augmentation to supplement datasets
- Synthetic data creation
- Image restoration
- Classifier on image patches
- Clinical records classification
The UNet model has achieved remarkable results in these applications, particularly in terms of the speed and accuracy of segmentation performance.
Also Read: Introduction to Autoencoders and Common Issues and Challenges
Conclusion
UNet is a powerful deep learning architecture that is widely used in image segmentation tasks. Its architecture is designed to preserve the spatial information of the original image while reducing the spatial resolution of the features. UNet is based on the encoder-decoder architecture and is trained using a combination of loss functions and datasets to produce a dense classification of the entire image. UNet’s applications in medical image analysis, remote sensing, and computer vision demonstrate its potential in real-life scenarios.
References
“What Is U-Net?” Educative: Interactive Courses for Software Developers, https://www.educative.io/answers/what-is-u-net. Accessed 6 Feb. 2023.
“Papers with Code – U-Net Explained.” Papers With Code, https://paperswithcode.com/method/u-net. Accessed 6 Feb. 2023.
Ronneberger, et al. “U-Net: Convolutional Networks for Biomedical Image Segmentation.” Springer International Publishing, 1 Jan. 2015, https://link.springer.com/chapter/10.1007/978-3-319-24574-4_28. Accessed 13 Feb. 2023.
Çiçek, et al. “3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation.” Springer International Publishing, 1 Jan. 2016, https://link.springer.com/chapter/10.1007/978-3-319-46723-8_49. Accessed 13 Feb. 2023.
Sha, Youyang, et al. “Transformer-Unet: Raw Image Processing with Unet.” arXiv.Org, 17 Sept. 2021, https://arxiv.org/abs/2109.08417. Accessed 13 Feb. 2023.
“UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation.” IEEE Xplore, https://ieeexplore.ieee.org/abstract/document/9053405/. Accessed 13 Feb. 2023.